Motivation: Discovering drug’s Anatomical Therapeutic Chemical (ATC) classification rules at molecular level is of vital importance to understand a vast majority of drugs action. However, few studies attempt to annotate drug’s potential ATC-codes by computational approaches.
Results: Here, we introduce drug-target network to computationally predict drug’s ATC-codes and propose a novel method named NetPredATC. Starting from the assumption that drugs with similar chemical structures or target proteins share common ATC-codes, our method, NetPredATC, aims to assign drug’s potential ATC-codes by integrating chemical structures and target proteins.
Specifically, we first construct a gold-standard positive dataset from drugs’ ATC-code annotation databases. Then we characterize ATC-code and drug by their similarity profiles and define kernel function to correlate them. Finally, we use a kernel method, support vector machine, to automatically predict drug’s ATC-codes. Our method was validated on four drug datasets with various target proteins, including enzymes, ion channels, G-protein couple receptors and nuclear receptors. We found that both drug’s chemical structure and target protein are predictive, and target protein information has better accuracy. Further integrating these two data sources revealed more experimentally validated ATC-codes for drugs. We extensively compared our NetPredATC with SuperPred, which is a chemical similarity-only based method. Experimental results showed that our NetPredATC outperforms SuperPred not only in predictive coverage but also in accuracy. In addition, database search and functional annotation analysis support that our novel predictions are worthy of future experimental validation.
Conclusion: In conclusion, our new method, NetPredATC, can predict drug’s ATC-codes more accurately by incorporating drug-target network and integrating data, which will promote drug mechanism understanding and drug repositioning and discovery.
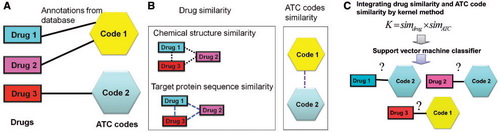
Fig. The scheme of our ATC-code prediction approach for drugs.
(A) Formulating known drug ATC-code annotations as a bipartite graph.
(B) Extracting drug–drug and ATC-code–ATC-code similarity metrics.
(C) Feeding the similarities among drugs and the similarities among ATC-codes to kernel method and applying SVM-based classifier to predict the unknown relationships between drugs and ATC-codes
1 Author Information:Yongcui Wang, Shilong Chen, Naiyang Deng and Yong Wang
Correspondence: E-mail:ycwang@nwipb.cas.cn or ywang@amss.ac.cn
2 Published:
Bioinformatics, 2013, 29:10, 1317-1324
