Rhubarb is a plant extensively utilized globally for both dietary and medicinal purposes. As wild resources become increasingly scarce, there is a growing interest in artificially cultivated rhubarb. The plant bacteriome is crucial for enhancing plant growth and metabolism. However, the composition and functions of plant bacteriome communities, under the influence of natural and anthropogenic factors, remain largely unexplored in wild and cultivated rhubarb. Here, we conducted 16S rRNA gene amplicon sequencing to focus on bacterial samples from bulk soil, rhizosphere soil, root, petiole, and leaf of wild and cultivated Rheum tanguticum at two sites with significant differences in altitude. Our findings indicated that, while host selection was the primary influencing factor, significant differences existed in the composition, function, community structure, and network complexity of the bacterial communities between cultivated and wild R. tanguticum. Cultivation activities altered the relative abundance of bacterial functional groups associated with the nitrogen cycle and significantly increased network complexity. However, these differences were mainly for root-associated bacteria but not for aboveground tissues. Differences in soil chemical properties resulting from fertilization may be the primary factor influencing the variation in bacterial communities between cultivated and wild soils. Structural equation model results also showed that changes in soil total nitrogen induced by fertilizer application directly affect the composition of root-associated bacteria. In addition, we also observed differences in the main active components of wild and cultivated R. tanguticum roots at different altitudes, which were significantly affected by the taxonomic composition and functional characteristics of the root-associated bacteria. This initial insight not only establishes a foundation for further exploration of the intricate interactions among bacteria, host plants, and environmental factors but also fosters the advancement of the R. tanguticum industry, thereby promoting its high-quality and sustainable development.
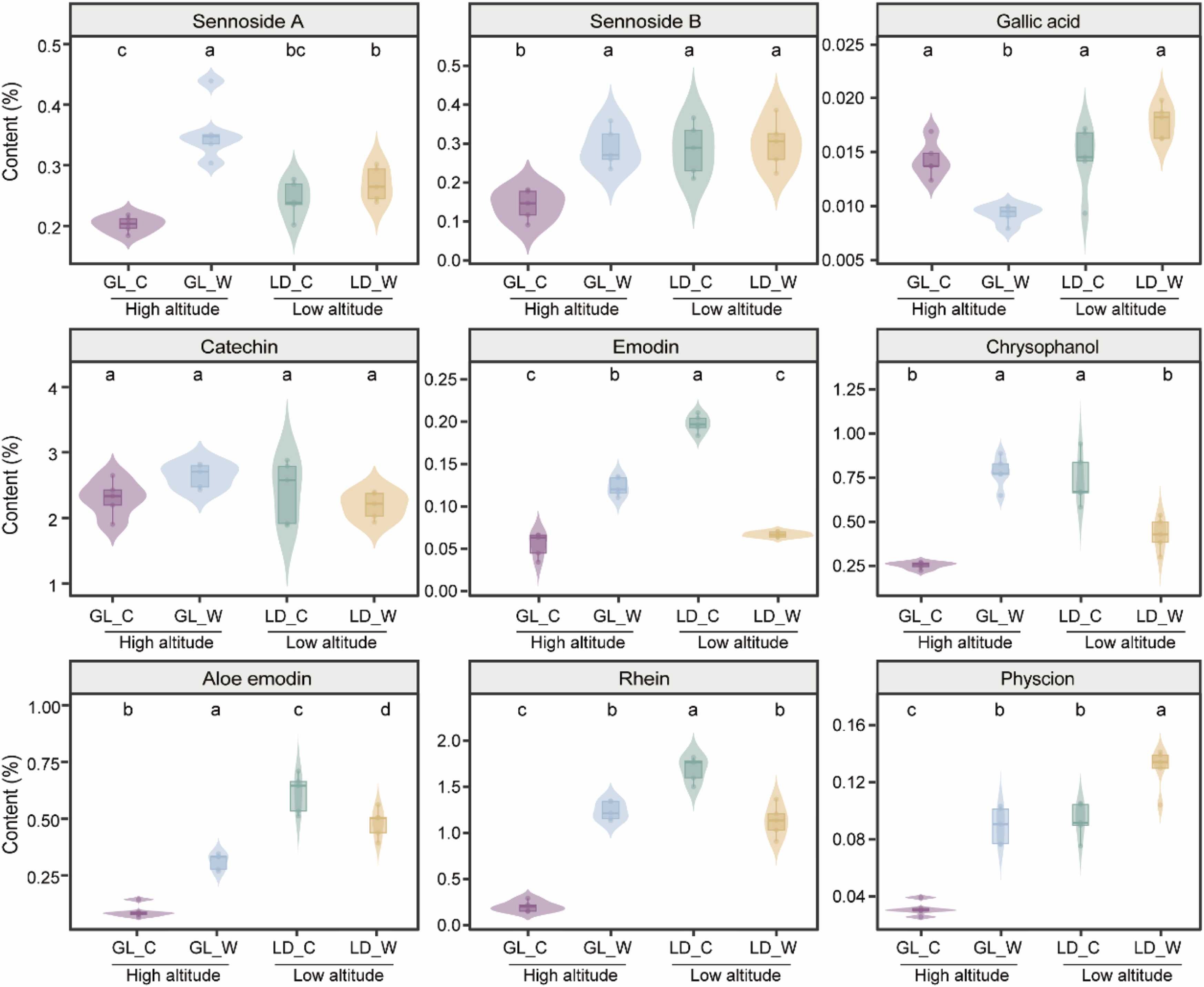
Fig. 1. Contents of active components in the roots of wild and cultivated R. tanguticum at different altitudes. Different letters indicated significant differences by Tukey’s HSD test. “GL_C” indicates the cultivated R. tanguticum in Guoluo. “GL_W” indicates the wild R. tanguticum in Guoluo. “LD_C” indicates the cultivated R. tanguticum in Ledu. “LD_W” indicates the wild R. tanguticum in Ledu.
The link below will guide you to the reading:
https://doi.org/10.1016/j.indcrop.2025.120657
