Accurate evaluation of fish stock biomass is essential for effective conservation management and targeted species enhancement efforts. However, this remains challenging owing to limited data availability. Therefore, we present an integrated modeling framework combining catch per unit effort with ensemble species distribution modeling called CPUESDM, which explicitly assesses and validates the spatial distribution of stock biomass for freshwaterfish species with limited data, applied to Herzensteinia microcephalus. The core algorithm incorporates the Leslie regression model, ensemble species distribution modeling, and exploratory spatial interpolation techniques. We found that H. microcephalus biomass in the Yangtze River source area yielded an initial estimate of 113.52 tons.
Our validation results demonstrate high accuracy with a Cohen's kappa coefficient of 0.78 and root mean square error of 0.05. Furthermore, our spatially-explicit, global, absolute biomass density map effectively identified areas with high and low concentrations of biomass distribution centers. Additionally, this study offers access to the source code, example raw data, and a step-by-step instruction manual for other researchers using field data to explore the application of this model. Our findings can help inform for future conservation efforts around fish stock biomass estimation, especially for endangered species.
The link below will guide you to the reading:
https://doi.org/10.1016/j.scitotenv.2023.168717
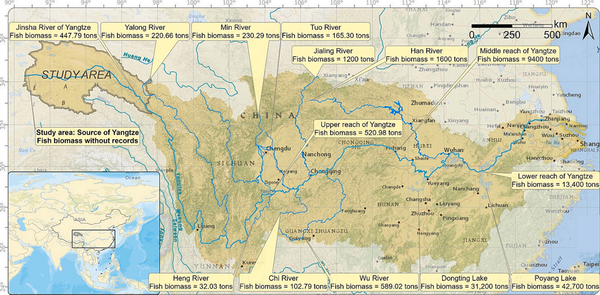
Topographic map displaying the location of the study area in the Yangtze Basin. The fish biomass in 2020 in the Yangtze and its tributaries and lakes (data from Dong et al., 2023 is marked by boxes in yellow
