Yak is one of the largest ruminants living in the areas with the highest average altitude in the world and physiologically adapted to hypoxic and cold environment. The domestic yak normally lives between 3000 and 5000m above sea level while the wild yak, the ancestor of domestic yak, inhabits at elevations from 4000 to 6000m on the QTP. Archaeological and molecular evidences revealed that wild yak was domesticated at least 7300 years ago. Although significant morphologic differences exist between wild and domestic yak1, both species share genetic features of high-altitude adaptation and are considered excellent models for studying hypoxia tolerance in large mammals.
In 2012, the genome of a female domestic yak was sequenced using the Illumina-based whole-genome shotgun method. In 2020, the genome of a female wild yak was assembled with the Illumina data, and chromosome-scale genomes of female domestic yak were constructed using long-read sequencing and chromatin interaction technologies. However, previous studies on yak have primarily focused on single nucleotide variants to reveal the genomic diversity and historical population dynamics. Recent studies showed that structural variants (SVs), such as insertions, deletions, duplications, inversions, and translocations, are widely present in genomes and provide an extensive source of genetic variations for identifying candidate genes involved in the regulation of critical biological processes. The availability of high-quality reference genomes for taurine cattle, which were obtained using long-read sequencing technologies, has enabled the dissection of genetic basis of complex traits. Assembling high-quality and complete reference genomes of wild and domestic yak is fundamental for deciphering the molecular mechanisms underpinning adaptation of yak and related species to extreme high-altitude environment on the QTP. Unfortunately, due to quality-related issues of the wild yak reference genome, SVs and SV-related genes in wild yak and domestic yak have yet to be mapped and compared with those in taurine cattle genome in detail.
Unique genomic features and precisely controlled gene expression ensure the physiological adaptation of animals to high altitude. Non-native mammals are prone to altitude sickness, mainly manifested as pulmonary hypertension and right ventricular hypertrophy after their exposure to acute or long-term hypoxia Yak acquired specific anatomical and physiological characteristics to survive the oxygen-poor air and the harsh environment. It is known that their blood, lung, and heart systems have been evolved to meet the challenge at high altitude23. Wild and domestic yak are adapted to hypoxia by increasing hemoglobin content, red blood cell count and hematocrit. The lung and heart weights of yak are higher than those of age-matched taurine cattle. Among these tissues, the lung is the interface between environment and body, therefore it plays a pivotal role in high-altitude adaptation. Yak developed intense pulmonary blood vessels, which increase the oxygen exchange rate of pulmonary artery blood vessels and help relieve pulmonary artery pressure. By comparing transcriptomic profiles among different tissues across species, it was reported that the lung exhibited adaptive transcriptional changes and expressed a higher number of positively selected genes. Despite these findings, the cellular components, and gene expression dynamics of lung tissues in animals that are adapted to high altitude remain to be explored at single cell level.
In the present study, we used Nanopore sequencing and Hi-C data to assemble the high-quality genomes of a wild yak and a domestic yak. We used an alignment-based strategy to compare long-read sequencing data with taurine cattle, to identify SVs associated with high altitude adaptation. We also constructed the single-cell atlas of yak and taurine cattle lungs using single-cell transcriptome sequencing (scRNA-seq). Integrated genomics and transcriptomics analyses revealed a new subtype of endothelial cells and uncovered a list of genes and pathways that were associated with the development of unique structures in yak lung. All together, these data provided important information to understand genetic and cellular mechanisms underlying the adaptive evolution of yak.
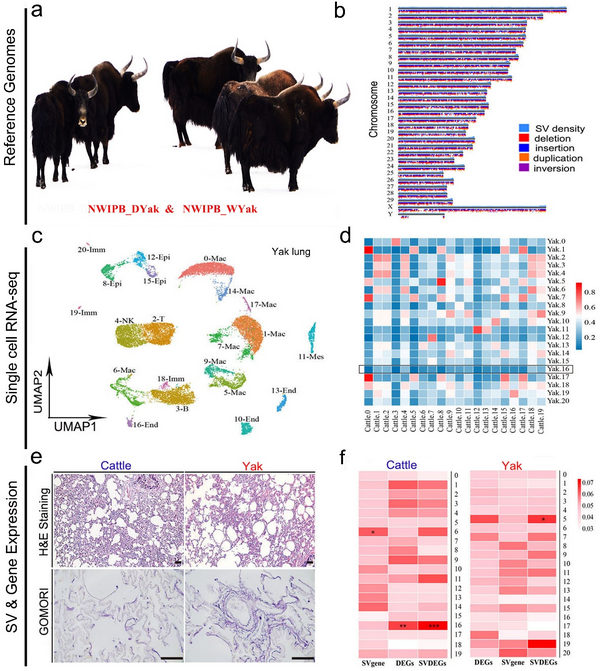
High quality reference genomes of Domestic Yak and wild yak were constructed, and the potential functions of genomic structural variation in gene expression regulation and lung cell differentiation were revealed combined with transcriptome, epigenome, single-cell transcriptome and histology
This result was published in Nature Communications with the title of “Long read genome assemblies complemented by single cell RNA-sequencing reveal genetic and cellular mechanisms underlying the adaptive evolution of yak". And it was reported by Science News with the title of "Yaks breathe easy thanks to newly discovered lung cells".
The link below will guide you to the reading:
https://www.nature.com/articles/s41467-022-32164-9
https://www.science.org/content/article/yaks-breathe-easy-thanks-newly-discovered-lung-cells
