Early flowering is a critical adaptive trait that strongly influences reproductive fitness in a short growing season. The circadian clock is a major regulator of photoperiod sensitivity, and mutations in circadian clock genes can be responsible for modifying the photoperiod sensitivity of may plant species, including barley (Hordeum vulgare). Loss-of-function mutations at the circadian clock gene EARLY MATURITY 8 (EAM8) promote rapid flowering. The early flowering of the EAM8 mutant mat.a-8 has been associated with an increased production of gibberellin, which cooperates with the product of FT1 to activate the expression of floral identity genes. Early flowering barleys incorporating the mat-a mutant have been successfully deployed in northern Scandinavia and Iceland where the growing season is short because of the low temperatures experienced in the spring and autumn. Early flowering cultivars have also been grown at high altitude regions close to the Equator to enable more than one crop per year to be grown.
SHEN Yuhu and his team identified a novel, spontaneous recessive eam8 mutant with an early flowering phenotype in a Tibetan barley landrace Lalu, which is natively grown at a high altitude of approximately 4000 m asl. The co-segregation analysis in a F2 population derived from the cross Lalu (early flowering) × Diqing 1 (late flowering) confirmed that early flowering of Lalu was determined to be due to an allele at EAM8. The eam8 allele from Lalu carries an A/G alternative splicing mutation at position 3257 in intron 3, designated eam8.l; this alternative splicing event leads to intron retention and a putative truncated protein. Of the 134 sequenced barley accessions, which are primarily native to the Qinghai-Tibet Plateau, three accessions carried this mutation. The eam8.l mutation was likely to have originated in wild barley due to the presence of the Lalu haplotype in H. spontaneum from Tibet. Overall, alternative splicing has contributed to the evolution of the barley circadian clock and in the short-season adaptation of local barley germplasm. The study also has identified a novel donor of early flowering barley which will be useful for barley improvement.
The relative study results were reported on Theoretical and Applied Genetics, a top journal of plant Genetics & Breeding, on March 2017. (http://link.springer.com/10.1007/s00122-016-2848-2)
Additional Information:
1 Author Information: Tengfei Xia, Lianquan Zhang, Jinqing Xu, Lei Wang, Baolong Liu, Ming Hao, Xi Chang , Tangwei Zhang, Shiming Li, Huaigang Zhang, Dengcai Liu*, and Yuhu Shen*.
Correspondence: shenyuhu@nwipb.cas.cn; dcliu7@yahoo.com.
2 Published: Theoretical and Applied Genetics, 2017, doi:10.1007/s00122-016-2848-2
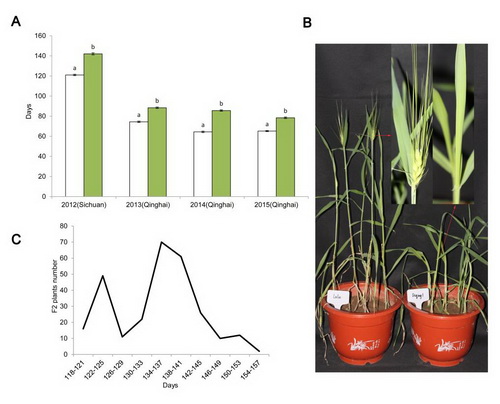
Fig. 1 Variation in the flowering time of Lalu and Diqing 1 plants grown in Sichuan and Qinghai. a Days to flowering (mean number of days to reach awn emergence on the leading tiller) of Lalu (white bars) and Diqing 1 (green bars). The error bars represent the standard error calculated from 19 Lalu plants and 13 Diqing 1 plants grown in Sichuan in 2012, and from 15 Lalu and 15 Diqing 1 plants grown in Qinghai in 2013-2015. b 60 day old plants of Lalu (left) and Diqing 1 (right). The arrows indicate the region of the leading tiller shown in magnified form. c The distribution of flowering time exhibited by the F2 progeny of the cross Lalu × Diqing 1.
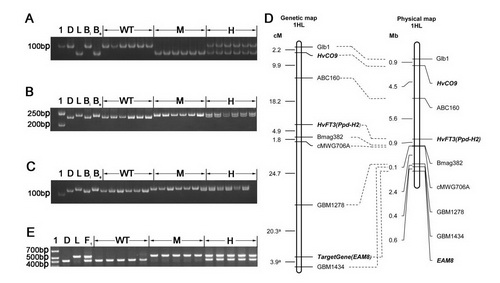
Fig. 2 Mapping the gene underlying early flowering in Lalu. Amplicons obtained from the SSR assays a Bmag382, b GBM1278 and c GBM1434. d A genetic (left) and a physical (right) map of the region of chromosome arm 1HL where the gene was located. The genetic map incorporates loci mapped by Kikuchi et al. (2012). e Amplicons obtained from a PCR assay based on the primer pair EAM8-2822-F/EAM8-3235-R, which targets the 104 bp indel in intron 2. 1: Molecular weight marker, D: Diqing1, L: Lalu, Be: a DNA bulk prepared from 20 early flowering F2 segregants; Bl: a DNA bulk prepared from 20 late flowering F2 segregants, WT: homozygote for the Diqing 1 allele, M: homozygote for the Lalu allele, H: heterozygote, F1: Lalu × Diqing 1 F1 plant.
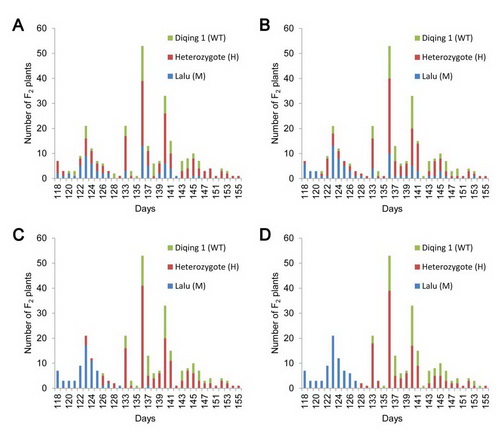
Fig. 3 Correlations between phenotype and genotype of a Bmag382, b GBM1278, c GBM1434 and d the 104 bp indel in intron 2 in F2 population. WT: homozygote for the Diqing 1 allele, M:
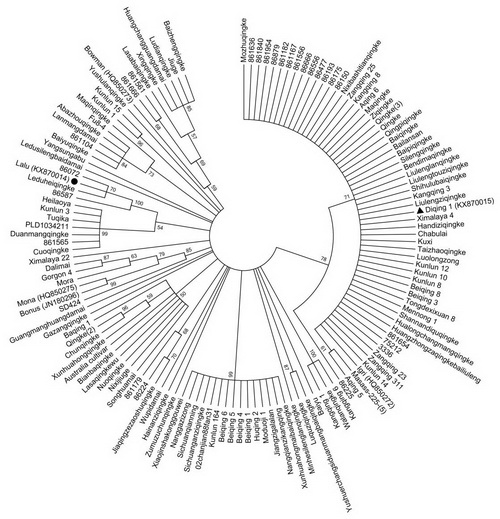
Fig. 4 Phylogenetic analysis of the haplotype germplasm panel based on the EAM8 sequences of 134 accessions. Lalu is indicated by a solid circle and Diqing 1 by a solid triangle. The bootstrap values are indicated near nodes.