Prof. Dou Quanwen and his team of Northwest Institute of Plateau Biology, the Chinese Academy of Sciences (NWIPB, CAS) identified eleven tandemly repetitive sequences from a Cot-1 library by FISH and sequence analysis of alfalfa (Medicago sativa), of which nine novel tandemly repetitive sequences can be adopted as distinct chromosome markers in alfalfa.
The researchers suggested a molecular karyotype of alfalfa with the chromosome markers they identified, and these novel chromosome markers will be a powerful tool for genome composition analysis, phylogenetic studies and breeding applications.
The study entitled “Cloning and characterization of chromosomal markers in alfalfa (Medicago sativa L.)” has been published in Theor Appl Genet.
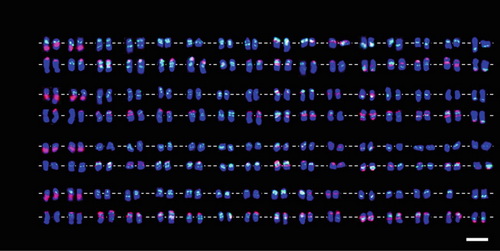
Fig. Karyotyping of four alfalfa cultivars with different FISH combinations.
T1, T2, T3, and T4 are cultivars ‘Zhongmu . 1’, ‘Aohan’, ‘Golden Empress’, and ‘Algonquin’, respectively.
a Probed by E180 (green) combined with 18S–26S rDNA (red) and 5S Rdna (red).
b Probed by E180 (green) combined with MsTR-1 (red) and MsCR-3 (red).
Scale bar = 10μm
Additional Information:
1 Author Information: Yu Feng, Lei Yunting, Li Yuan, Dou Quanwen, Wang Haiqing, Chen Zhiguo
Correspondence: douqw@nwipb.cas.cn.
2 Published:
Theor Appl Genet (2013) 126:1885–1896
