Wild Triticeae grasses serve as important gene pools for forage and cereal crops. Based on DNA sequences of genomespecific RAPD markers, sequence-tagged site (STS) markers specific for W and Y genomes have been obtained. Coupling with the use of genomic in situ hybridization, these STS markers enabled the identification of the W- and Y-genome chromosomes in backcross derivatives from hybrids of bread wheat Triticum aestivum L. (2n = 42; AABBDD) and Elymus rectisetus (Nees in Lehm.) Á. Löve & Connor (2n = 42; StStWWYY). The detection of six different alien chromosomes in five of these derivatives was ascertained by quantitative PCR of STS markers, simple sequence repeat markers, rDNA genes, and (or) multicolor florescence in situ hybridization. Disomic addition line 4687 (2n = 44) has the full complement of 42 wheat chromosomes and a pair of 1Y chromosomes that carry genes for resistance to tan spot (caused by Pyrenophora tritici-repentis (Died.) Drechs.) and Stagonospora nodorum blotch (caused by Stagonospora nodorum (Berk.) Castellani and Germano). The disomic addition line 4162 has a pair of 1St chromosomes and 21 pairs of wheat chromosomes. Lines 4319 and 5899 are two triple substitution lines (2n = 42) having the same chromosome composition, with 2A, 4B, and 6D of wheat substituted by one pair of W- and two pairs of St-genome chromosomes. Line 4434 is a substitution–addition line (2n = 44) that has the same W- and St-genome chromosomes substituting 2A, 4B, and 6D of wheat as in lines 4319 and 5899 but differs by having an additional pair of Y-genome chromosome, which is not the 1Y as in line 4687. The production and identification of these alien cytogenetic stocks may help locate and isolate genes for useful agronomic traits. Key words: addition line, substitution line, genome, apomixis, disease resistance.
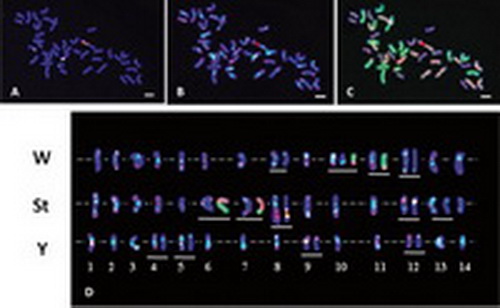
Fig. Mitotic chromosomes in Elymus rectisetus (PI 533028) analyzed by sequential FISH and GISH techniques.
(A) Chromosomes probed by 5S rDNA (labeled with fluorescein-12-dUTP for green color) and 45S rDNA (labeled with tetramethy1-rhodamine-5-dUTP for red color).
(B) Probed by pAs1 (red) and (AAG)10 microsatellite sequence (green).
(C) Probed by W-genomic DNA (green) and St-genomic DNA (red).
(D) FISH karyotype of W-, St-, and Y-genome chromosomes arranged according to length and arm ratios.
Chromosomes having rRNA genes or intergenomic translocations are underlined. Note that the 14 chromosomes of each genome could not be paired into seven homologous groups.

Fig. Alien chromosomes, originated from the W, St, or Y genome of Elymus rectisetus, in the backcross derivatives of Triticum aestivum × E. rectisetus hybrids.
Lines 4687 and 4162 possess 1Y and 1St that has the 45S and 5S rDNA, respectively.
Lines 4319, 5899, and 4434 might share the same one pair W- and two pairs of St-genome chromosomes.
mW is W-14, nSt is St-4, pSt is St-3, and qY is Y-2 in Fig. 5D. 1Y and 1St are corresponding to W-9 and St-13 in Fig. 5D, respectively.
Additional Information:
1 Author Information: Quan-Wen Dou, Yunting Lei, Xiaomei Li, Ivan W. Mott, and Richard R.-C. Wang.
Correspondence: Richard R.-C. Wang (e-mail: Richard.Wang@ars.usda.gov).
2 Published:
Genome, 2012, 55(5): 337-347, 10.1139/g2012-018
Published on the web 11 April 2012.
1 Author Information: Quan-Wen Dou, Yunting Lei, Xiaomei Li, Ivan W. Mott, and Richard R.-C. Wang.
Correspondence: Richard R.-C. Wang (e-mail: Richard.Wang@ars.usda.gov).
2 Published:
Genome, 2012, 55(5): 337-347, 10.1139/g2012-018
Published on the web 11 April 2012.
