Lycium ruthenicum (black goji) is a medicinal plant native to the Qinghai-Tibet Plateau (Cao et al., 2021), known for its high anthocyanin content (Avula et al., 2023) in fruit. In contrast, the white and purple variants contain little anthocyanin (Zong et al., 2019). The evolutionary relationship of the variants and the genetic basis underlying their color differentiation has rarely been well studied at the whole genome level (Li et al., 2024). In this study, we present a near-complete genome assembly of L. ruthenicum, providing a valuable resource for investigating its evolutionary relationships with other Lycium species and fruit color variants. Through integrated genomic, transcriptomic, and functional analyses, we identify a key structural variation of AN1, a bHLH transcription factor essential for anthocyanin biosynthesis, which underlies the formation of white and purple goji in L. ruthenicum.
A high-quality genome of L. ruthenicum cultivar ‘Qingheiqi1hao’ (QHQ1) was assembled using 308 Gb of PacBio SMRT, 84.8 Gb of PacBio Revio HiFi, and 92 Gb of Illumina paired-end (PE150) reads, resulting in a 2.37 Gb genome with a contig N50 of 107.9 Mb. With the aid of 436 Gb of Hi-C data, 2.22 Gb (99.1%) of the genome was anchored to 12 pseudochromosomes (Fig. 1, A; Fig. S1), consistent with the karyotype analysis (Fig. S2). Genome sequencing data have been deposited in NCBI (PRJNA1152636). Genome assemblies and annotations are available at Figshare (https://doi.org/10.6084/m9.figshare.26830252.v1). Two chromosomes were assembled at telomere-to-telomere resolution. Approximately 79.8% of the genome was annotated as repetitive sequences, with LTR-Gypsy elements accounting for the majority (54.6%). In total, 17 telomeres and 10 centromeres were identified based on specific repeat sequences (Fig. 1, B). A total of 36 434 high-confidence (HC) genes were predicted, of which 97.65% (35 579) were annotated in the NR, KEGG, Swiss-Prot, and TrEMBL databases. BUSCO analysis based on protein sequences indicated an overall gene set completeness of 95.6%. Comparative analysis with the recently published L. ruthenicum genome (ASM4143038v1) revealed high collinearity across most chromosomes, except for chromosome 2, which exhibited a higher LTR content in the QHQ1 assembly (58.9% vs 50.7%) (Fig. S3). Moreover, the QHQ1 genome demonstrated greater assembly contiguity (N50 = 107.9 Mb vs 92.6 Mb) and more comprehensive KEGG annotation (27 718 vs 9 304 genes), reflecting superior assembly quality and functional completeness.
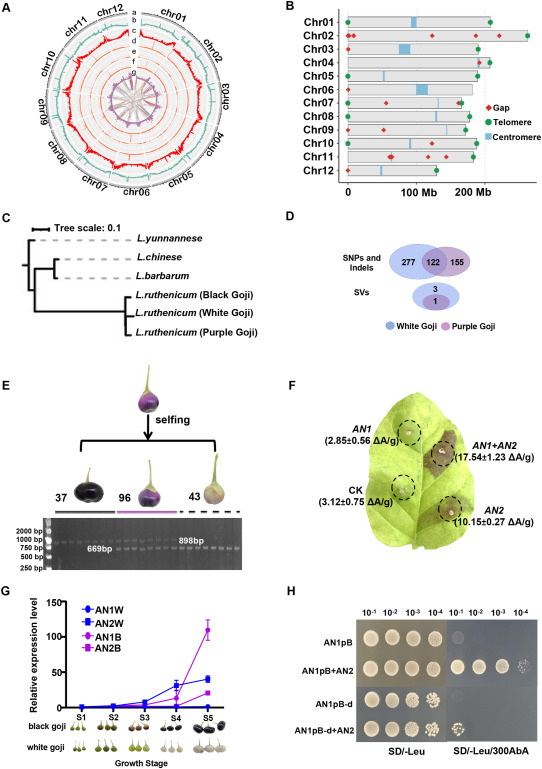
Fig. 1. High-quality genome assembly and functional dissection of a 229-bp AN1 promoter deletion underlying fruit color variation in L. ruthenicum
The link below will guide you to the reading:
http://english.nwipb.cas.cn/ns/rn/202508/t20250822_1051265.html
